AutoCellLabel Live
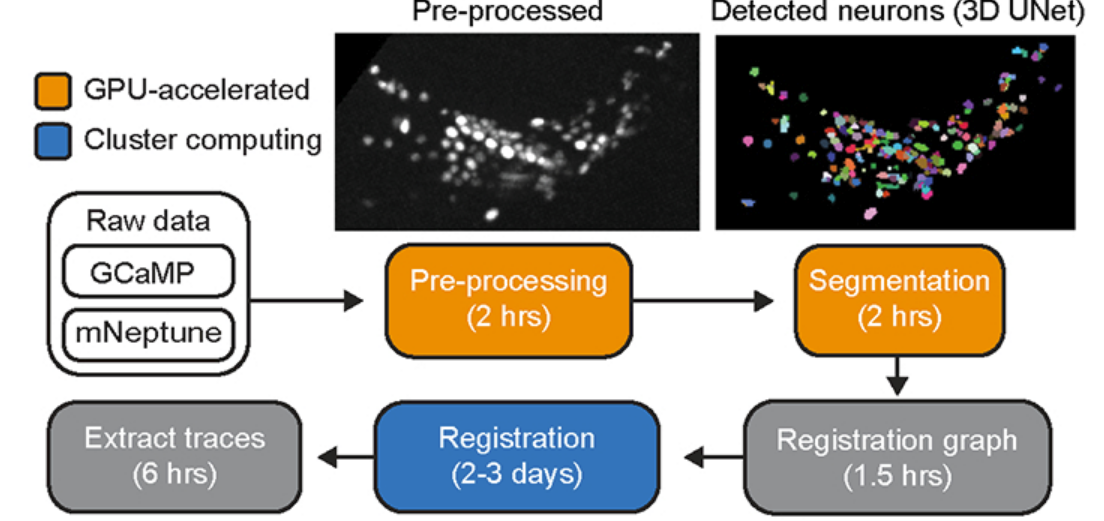
If we wanted to get real time labeling working, the previous approach would simply not work. First, BrainAlignNet was a non-starter, and the computational difficulty of the non-rigid registration problem was not tractable to perform online. Secondly, we couldn't wait until the end of a recording to get our labels, and would need to move past immobilization, instead performing accurate inference on freely moving frames. Luckily, these problems lend themselves to an elegant mutual solution: if we can label each frame accurately, we no long need to match neurons across freely moving frames. (and, we can still use BrainAlignNet post-doc to perform ANTSUN labeling for verification or even just aggregate freely moving predictions and pick up more neurons than standard ANTSUN).
From that idea, I began working on AutoCellLabel Live. ACLL is a pipeline, mostly focused on the FreelyMoving AutoCellLabeler network, but also encompassing highly accelerated preprocessing and trace extraction procedures. Unfortunately, recording practicalities mean we only have one low-SNR fluorescence channel to work with, instead of four high-SNR channels in the immobilized images. Luckily, we also have 1600 times the number of volumes to train on, as each recording only contains one immobilized image. This, however, meant ACL training would have to speed up, as the published version took more than a week to train on just 81 frames. However, even once the network existed, its inference time would need to be drastically decreased, as it look just over a minute to run. We record a volume every 0.6 seconds, so the entire pipeline would need to complete in that time.
Straight Efficiency Gains
- Heavy rewriting of the network and loss, optimization of the data loader, and access to better hardware allowed for faster training (from a week on 81 training frames to 5 hrs on 8100 frames)
- Intensive tracing focused on code and data management optimization resulted in faster inference (1 minute -> 0.91 seconds)
- Significantly decreased peak memory usage in training and inference allows for batching in both
- Custom NIS Elements acquisition setup to ensure data can be accessed and processed in real time (this was a pain)
Pipeline Refactoring Gains
- Shear correction was prohibitive by itself (>1 second) but turns out you can train a UNet to perform shear correction... so we trained FM ACL to predict on sheared data, eliminating the need for a separate shear-correction step
- Channel alignment (xy offset of signal channel vs reference labeling channel) was likewise prohibitive (2.5 seconds) but is relatively consistent across each recording. We can take a small sample of the first few frames, do the heavy computation to determine the offset, and apply that to the rest off the frames, which is much faster
- Naive ROI extraction from ACL predictions removed the need for a segmentation network
Results
- After 5 frames to establish our channel alignment parameters, the pipeline runs in 1.2 seconds, which allows us to meet our required FPS with a batch size of 2.
- Unfortunately, traces are noticeably noisier, and sometimes completely unlike those produced by ANTSUN; the lack of proper segmentation means extraneous pixels can sneak in.
- However, performance is consistent across a recording (i.e. if a trace starts out looking good, it will keep looking good). Performance is also well correlated with accuracy, which is well correlated with confidence. Meaning we can quickly determine which traces are likely to be good in a recording.